Deep Learning básico con Keras (Parte 3): VGG
En este tercer artículo vamos a mostrar la red VGG. Introducida en el siguiente paper, Very Deep Convolutional Networks for Large-Scale Image Recognition, Es una de las primeras redes profundas más conocidas. Os muestro en las dos imágenes siguientes el modelo y sus especificaciones.
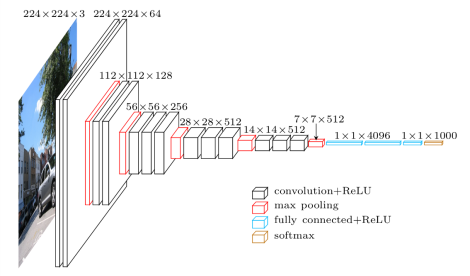
Diagrama de arquitectura general VGG
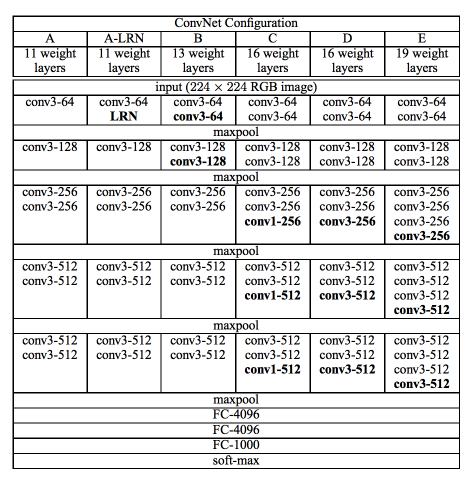
Arquitecturas VGG
Realizaremos el mismo experimento que en las partes anteriores. Obviaremos los puntos en los que importamos el dataset de CIFAR-100, la configuración básica del entorno del experimento y la importación de las librerías de python, pues son exactamente igual.
Entrenando la arquitectura VGG-16
Keras tiene a nuestra disposición tanto la arquitectura VGG-16 como la VGG-19. Vamos a entrenar ambas y una más que explicaremos en breve. Debido a que vamos a usar la definida en Keras (aunque podríamos crearla directamente), debemos aumentar el tamaño de las imágenes a 48 píxeles. Para ello, usaremos el siguiente código:
def resize_data(data):
data_upscaled = np.zeros((data.shape[0], 48, 48, 3))
for i, img in enumerate(data):
large_img = cv2.resize(img, dsize=(48, 48), interpolation=cv2.INTER_CUBIC)
data_upscaled[i] = large_img
return data_upscaled
x_train_resized = resize_data(x_train_original)
x_test_resized = resize_data(x_test_original)
x_train_resized = x_train_resized / 255
x_test_resized = x_test_resized / 255
Con esto, tenemos las imágenes a 48 píxeles normalizadas en x_train_resized y x_test_resized. Como dijimos antes, entrenaremos VGG-16, VGG-19 y una más. Esta es nuestra propia versión de VGG con objeto de no tener que modificar el tamaño de las imágenes. Empezaremos entrenando VGG-16, veremos resultados y continuaremos con VGG-19 para terminar con nuestra Custom VGG.
VGG-16
Definimos el modelo. Tan fácil como llamar al existente en Keras.
from keras.applications import vgg16
def create_vgg16():
model = vgg16.VGG16(include_top=True, weights=None, input_tensor=None, input_shape=(48,48,3), pooling=None, classes=100)
return model
Los parámetros son sencillos, vamos a incluir una red neuronal densa al final con el parámetro include_top. No cargamos ningún modelo entrenado a priori con el parámetro weights. No especificamos ningún tensor de keras como entrada con input_tensor. Definimos la forma de los datos de entrada con input_shape, No especificamos Pooling final con pooling y definimos el número de clases final con classes.
Una vez definido el modelo, lo compilamos especificando la función de optimización, la de coste o pérdida y las métricas que usaremos. En este caso, como en los artículos anteriores, usaremos la función de optimización stochactic gradient descent, la función de pérdida categorical cross entropy y, para las métricas, accuracy y mse (media de los errores cuadráticos).
vgg16_model = create_vgg16()
vgg16_model.compile(loss='categorical_crossentropy', optimizer='sgd', metrics=['acc', 'mse'])
Una vez hecho esto, vamos a ver un resumen del modelo creado.
vgg16_model.summary()
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input_1 (InputLayer) (None, 48, 48, 3) 0
_________________________________________________________________
block1_conv1 (Conv2D) (None, 48, 48, 64) 1792
_________________________________________________________________
block1_conv2 (Conv2D) (None, 48, 48, 64) 36928
_________________________________________________________________
block1_pool (MaxPooling2D) (None, 24, 24, 64) 0
_________________________________________________________________
block2_conv1 (Conv2D) (None, 24, 24, 128) 73856
_________________________________________________________________
block2_conv2 (Conv2D) (None, 24, 24, 128) 147584
_________________________________________________________________
block2_pool (MaxPooling2D) (None, 12, 12, 128) 0
_________________________________________________________________
block3_conv1 (Conv2D) (None, 12, 12, 256) 295168
_________________________________________________________________
block3_conv2 (Conv2D) (None, 12, 12, 256) 590080
_________________________________________________________________
block3_conv3 (Conv2D) (None, 12, 12, 256) 590080
_________________________________________________________________
block3_pool (MaxPooling2D) (None, 6, 6, 256) 0
_________________________________________________________________
block4_conv1 (Conv2D) (None, 6, 6, 512) 1180160
_________________________________________________________________
block4_conv2 (Conv2D) (None, 6, 6, 512) 2359808
_________________________________________________________________
block4_conv3 (Conv2D) (None, 6, 6, 512) 2359808
_________________________________________________________________
block4_pool (MaxPooling2D) (None, 3, 3, 512) 0
_________________________________________________________________
block5_conv1 (Conv2D) (None, 3, 3, 512) 2359808
_________________________________________________________________
block5_conv2 (Conv2D) (None, 3, 3, 512) 2359808
_________________________________________________________________
block5_conv3 (Conv2D) (None, 3, 3, 512) 2359808
_________________________________________________________________
block5_pool (MaxPooling2D) (None, 1, 1, 512) 0
_________________________________________________________________
flatten (Flatten) (None, 512) 0
_________________________________________________________________
fc1 (Dense) (None, 4096) 2101248
_________________________________________________________________
fc2 (Dense) (None, 4096) 16781312
_________________________________________________________________
predictions (Dense) (None, 100) 409700
=================================================================
Total params: 34,006,948
Trainable params: 34,006,948
Non-trainable params: 0
_________________________________________________________________
Ahora el número de parámetros ha crecido sustancialmente (34 millones). Ahora sólo queda entrenar.
vgg16 = vgg16_model.fit(x=x_train_resized, y=y_train, batch_size=32, epochs=10, verbose=1, validation_data=(x_test_resized, y_test), shuffle=True)
Train on 50000 samples, validate on 10000 samples
Epoch 1/10
50000/50000 [==============================] - 173s 3ms/step - loss: 4.6053 - acc: 0.0095 - mean_squared_error: 0.0099 - val_loss: 4.6049 - val_acc: 0.0164 - val_mean_squared_error: 0.0099
Epoch 2/10
50000/50000 [==============================] - 171s 3ms/step - loss: 4.6050 - acc: 0.0114 - mean_squared_error: 0.0099 - val_loss: 4.6045 - val_acc: 0.0160 - val_mean_squared_error: 0.0099
Epoch 3/10
50000/50000 [==============================] - 171s 3ms/step - loss: 4.6041 - acc: 0.0158 - mean_squared_error: 0.0099 - val_loss: 4.6028 - val_acc: 0.0200 - val_mean_squared_error: 0.0099
Epoch 4/10
50000/50000 [==============================] - 171s 3ms/step - loss: 4.6001 - acc: 0.0185 - mean_squared_error: 0.0099 - val_loss: 4.5940 - val_acc: 0.0110 - val_mean_squared_error: 0.0099
Epoch 5/10
50000/50000 [==============================] - 171s 3ms/step - loss: 4.4615 - acc: 0.0278 - mean_squared_error: 0.0099 - val_loss: 4.3003 - val_acc: 0.0519 - val_mean_squared_error: 0.0098
Epoch 6/10
50000/50000 [==============================] - 171s 3ms/step - loss: 4.1622 - acc: 0.0687 - mean_squared_error: 0.0097 - val_loss: 4.1022 - val_acc: 0.0765 - val_mean_squared_error: 0.0097
Epoch 7/10
50000/50000 [==============================] - 171s 3ms/step - loss: 4.0242 - acc: 0.0888 - mean_squared_error: 0.0097 - val_loss: 4.0127 - val_acc: 0.0939 - val_mean_squared_error: 0.0097
Epoch 8/10
50000/50000 [==============================] - 171s 3ms/step - loss: 3.9118 - acc: 0.1052 - mean_squared_error: 0.0096 - val_loss: 4.0327 - val_acc: 0.0963 - val_mean_squared_error: 0.0096
Epoch 9/10
50000/50000 [==============================] - 171s 3ms/step - loss: 3.7884 - acc: 0.1235 - mean_squared_error: 0.0095 - val_loss: 3.7928 - val_acc: 0.1276 - val_mean_squared_error: 0.0094
Epoch 10/10
50000/50000 [==============================] - 171s 3ms/step - loss: 3.6518 - acc: 0.1429 - mean_squared_error: 0.0094 - val_loss: 3.8205 - val_acc: 0.1316 - val_mean_squared_error: 0.0095
Obviaremos la evaluación.
Veamos las métricas obtenidas para el entrenamiento y validación gráficamente.
plt.figure(0)
plt.plot(vgg16.history['acc'],'r')
plt.plot(vgg16.history['val_acc'],'g')
plt.xticks(np.arange(0, 11, 2.0))
plt.rcParams['figure.figsize'] = (8, 6)
plt.xlabel("Num of Epochs")
plt.ylabel("Accuracy")
plt.title("Training Accuracy vs Validation Accuracy")
plt.legend(['train','validation'])
plt.figure(1)
plt.plot(vgg16.history['loss'],'r')
plt.plot(vgg16.history['val_loss'],'g')
plt.xticks(np.arange(0, 11, 2.0))
plt.rcParams['figure.figsize'] = (8, 6)
plt.xlabel("Num of Epochs")
plt.ylabel("Loss")
plt.title("Training Loss vs Validation Loss")
plt.legend(['train','validation'])
plt.show()
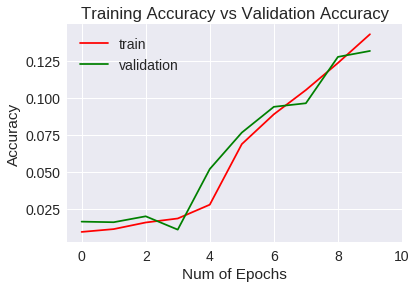
Accuracy
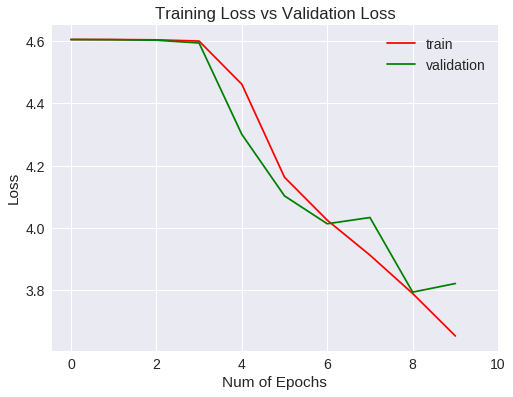
Loss
La generalización mejora a un 1% aproximadamente respecto a la red convolucional del experimento anterior, si bien, después de 10 epochs no ha mejorado las métricas estándar.
Matriz de confusión
Pasemos ahora a ver la matriz de confusión y las métricas de Accuracy, Recall y F1-score.
Vamos a hacer una predicción sobre el dataset de validación y, a partir de ésta, generamos la matriz de confusión y mostramos las métricas mencionadas anteriormente.
vgg16_pred = vgg16_model.predict(x_test_resized, batch_size=32, verbose=1)
vgg16_predicted = np.argmax(vgg16_pred, axis=1)
Como ya hiciéramos en la primera parte, vamos a dar como predecida el mayor valor de la predicción. Lo normal es dar un valor mínimo o bias que defina un resultado como positivo, pero en este caso, lo vamos a hacer simple.
Con la librería Scikit Learn, generamos la matriz de confusión y la dibujamos (aunque el gráfico no es muy bueno debido al numero de etiquetas).
#Creamos la matriz de confusión
vgg16_cm = confusion_matrix(np.argmax(y_test, axis=1), vgg16_predicted)
# Visualiamos la matriz de confusión
vgg16_df_cm = pd.DataFrame(vgg16_cm, range(100), range(100))
plt.figure(figsize = (20,14))
sn.set(font_scale=1.4) #for label size
sn.heatmap(vgg16_df_cm, annot=True, annot_kws={"size": 12}) # font size
plt.show()
Matriz de confusión
Y por último, mostramos las métricas
vgg16_report = classification_report(np.argmax(y_test, axis=1), vgg16_predicted)
print(vgg16_report)
precision recall f1-score support
0 0.63 0.27 0.38 100
1 0.10 0.14 0.11 100
2 0.04 0.04 0.04 100
3 0.01 0.01 0.01 100
4 0.02 0.01 0.01 100
5 0.05 0.04 0.05 100
6 0.16 0.03 0.05 100
7 0.11 0.11 0.11 100
8 0.06 0.01 0.02 100
9 0.31 0.18 0.23 100
10 0.14 0.18 0.15 100
11 0.20 0.01 0.02 100
12 0.11 0.31 0.16 100
13 0.09 0.26 0.13 100
14 0.13 0.10 0.11 100
15 0.00 0.00 0.00 100
16 0.16 0.25 0.19 100
17 0.17 0.40 0.24 100
18 0.00 0.00 0.00 100
19 0.09 0.03 0.04 100
20 0.52 0.25 0.34 100
21 0.06 0.07 0.07 100
22 0.21 0.20 0.21 100
23 0.14 0.38 0.21 100
24 0.28 0.30 0.29 100
25 0.07 0.09 0.08 100
26 0.10 0.03 0.05 100
27 0.07 0.03 0.04 100
28 0.35 0.24 0.28 100
29 0.06 0.14 0.09 100
30 0.12 0.55 0.20 100
31 0.08 0.08 0.08 100
32 0.30 0.06 0.10 100
33 0.17 0.05 0.08 100
34 0.06 0.05 0.06 100
35 0.06 0.04 0.05 100
36 0.06 0.02 0.03 100
37 0.08 0.14 0.10 100
38 0.04 0.01 0.02 100
39 0.08 0.08 0.08 100
40 0.27 0.21 0.24 100
41 0.50 0.45 0.47 100
42 0.05 0.09 0.07 100
43 0.15 0.06 0.09 100
44 0.07 0.01 0.02 100
45 0.00 0.00 0.00 100
46 0.08 0.01 0.02 100
47 0.20 0.11 0.14 100
48 0.17 0.11 0.13 100
49 0.29 0.07 0.11 100
50 0.00 0.00 0.00 100
51 0.16 0.03 0.05 100
52 0.14 0.85 0.24 100
53 0.23 0.11 0.15 100
54 0.25 0.03 0.05 100
55 0.09 0.02 0.03 100
56 0.09 0.30 0.14 100
57 0.33 0.08 0.13 100
58 0.03 0.01 0.01 100
59 0.00 0.00 0.00 100
60 0.56 0.51 0.53 100
61 0.28 0.28 0.28 100
62 0.18 0.34 0.23 100
63 0.08 0.05 0.06 100
64 0.10 0.03 0.05 100
65 0.08 0.02 0.03 100
66 0.02 0.01 0.01 100
67 0.03 0.01 0.01 100
68 0.29 0.58 0.39 100
69 0.18 0.09 0.12 100
70 0.00 0.00 0.00 100
71 0.27 0.47 0.34 100
72 0.10 0.16 0.12 100
73 0.33 0.02 0.04 100
74 0.00 0.00 0.00 100
75 0.10 0.40 0.16 100
76 0.14 0.37 0.20 100
77 0.05 0.01 0.02 100
78 0.00 0.00 0.00 100
79 0.00 0.00 0.00 100
80 0.03 0.01 0.01 100
81 0.05 0.10 0.06 100
82 0.25 0.23 0.24 100
83 0.43 0.06 0.11 100
84 0.13 0.02 0.03 100
85 0.17 0.10 0.13 100
86 0.23 0.13 0.17 100
87 0.08 0.01 0.02 100
88 0.03 0.03 0.03 100
89 0.17 0.06 0.09 100
90 0.14 0.02 0.04 100
91 0.12 0.38 0.18 100
92 0.00 0.00 0.00 100
93 0.05 0.09 0.06 100
94 0.19 0.16 0.17 100
95 0.09 0.39 0.15 100
96 0.06 0.01 0.02 100
97 0.04 0.18 0.07 100
98 0.04 0.02 0.03 100
99 0.11 0.07 0.09 100
avg / total 0.14 0.13 0.11 10000
Curva ROC (tasas de verdaderos positivos y falsos positivos)
Vamos a codificar la curva ROC para clasificación multiclase. Como hemos dicho en los artículos anteriores, el código está obtenido del blog de DloLogy, pero se puede obtener de la documentación de Scikit Learn.
from sklearn.datasets import make_classification
from sklearn.preprocessing import label_binarize
from scipy import interp
from itertools import cycle
n_classes = 100
from sklearn.metrics import roc_curve, auc
# Plot linewidth.
lw = 2
# Compute ROC curve and ROC area for each class
fpr = dict()
tpr = dict()
roc_auc = dict()
for i in range(n_classes):
fpr[i], tpr[i], _ = roc_curve(y_test[:, i], vgg16_pred[:, i])
roc_auc[i] = auc(fpr[i], tpr[i])
# Compute micro-average ROC curve and ROC area
fpr["micro"], tpr["micro"], _ = roc_curve(y_test.ravel(), vgg16_pred.ravel())
roc_auc["micro"] = auc(fpr["micro"], tpr["micro"])
# Compute macro-average ROC curve and ROC area
# First aggregate all false positive rates
all_fpr = np.unique(np.concatenate([fpr[i] for i in range(n_classes)]))
# Then interpolate all ROC curves at this points
mean_tpr = np.zeros_like(all_fpr)
for i in range(n_classes):
mean_tpr += interp(all_fpr, fpr[i], tpr[i])
# Finally average it and compute AUC
mean_tpr /= n_classes
fpr["macro"] = all_fpr
tpr["macro"] = mean_tpr
roc_auc["macro"] = auc(fpr["macro"], tpr["macro"])
# Plot all ROC curves
plt.figure(1)
plt.plot(fpr["micro"], tpr["micro"],
label='micro-average ROC curve (area = {0:0.2f})'
''.format(roc_auc["micro"]),
color='deeppink', linestyle=':', linewidth=4)
plt.plot(fpr["macro"], tpr["macro"],
label='macro-average ROC curve (area = {0:0.2f})'
''.format(roc_auc["macro"]),
color='navy', linestyle=':', linewidth=4)
colors = cycle(['aqua', 'darkorange', 'cornflowerblue'])
for i, color in zip(range(n_classes-97), colors):
plt.plot(fpr[i], tpr[i], color=color, lw=lw,
label='ROC curve of class {0} (area = {1:0.2f})'
''.format(i, roc_auc[i]))
plt.plot([0, 1], [0, 1], 'k--', lw=lw)
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Some extension of Receiver operating characteristic to multi-class')
plt.legend(loc="lower right")
plt.show()
# Zoom in view of the upper left corner.
plt.figure(2)
plt.xlim(0, 0.2)
plt.ylim(0.8, 1)
plt.plot(fpr["micro"], tpr["micro"],
label='micro-average ROC curve (area = {0:0.2f})'
''.format(roc_auc["micro"]),
color='deeppink', linestyle=':', linewidth=4)
plt.plot(fpr["macro"], tpr["macro"],
label='macro-average ROC curve (area = {0:0.2f})'
''.format(roc_auc["macro"]),
color='navy', linestyle=':', linewidth=4)
colors = cycle(['aqua', 'darkorange', 'cornflowerblue'])
for i, color in zip(range(3), colors):
plt.plot(fpr[i], tpr[i], color=color, lw=lw,
label='ROC curve of class {0} (area = {1:0.2f})'
''.format(i, roc_auc[i]))
plt.plot([0, 1], [0, 1], 'k--', lw=lw)
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Some extension of Receiver operating characteristic to multi-class')
plt.legend(loc="lower right")
plt.show()
El resultado para tres clases se muestra en los siguientes gráficos.
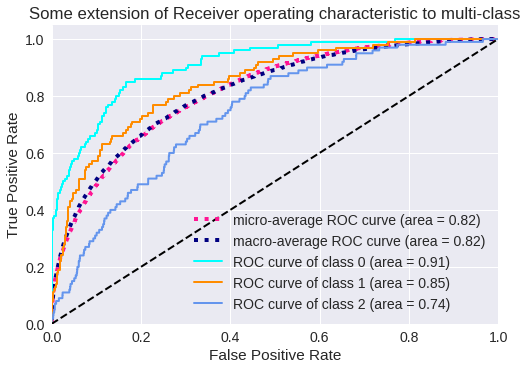
Curva ROC para 3 clases
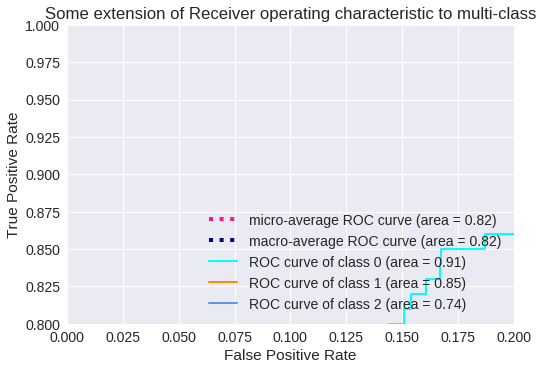
Zoom de la Curva ROC para 3 clases
Vamos a ver algunos resultados
imgplot = plt.imshow(x_train_original[0])
plt.show()
print('class for image 1: ' + str(np.argmax(y_test[0])))
print('predicted: ' + str(vgg16_predicted[0]))
Una vaca?
class for image 1: 49
predicted: 95
imgplot = plt.imshow(x_train_original[3])
plt.show()
print('class for image 3: ' + str(np.argmax(y_test[3])))
print('predicted: ' + str(vgg16_predicted[3]))

Un hombre
class for image 3: 51
predicted: 75
Salvaremos los datos del histórico de entrenamiento para compararlos con otros modelos.
#Histórico
with open(path_base + '/vgg16_history.txt', 'wb') as file_pi:
pickle.dump(scnn.history, file_pi)
VGG-19
Definamos el modelo y entrenemos de la misma forma que VGG-16.
def create_vgg19():
model = vgg19.VGG19(include_top=True, weights=None, input_tensor=None, input_shape=(48,48,3), pooling=None, classes=100)
return model
vgg19_model = create_vgg19()
vgg19_model.compile(loss='categorical_crossentropy', optimizer='sgd', metrics=['acc', 'mse'])
Una vez hecho esto, vamos a ver un resumen del modelo creado.
vgg19_model.summary()
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input_2 (InputLayer) (None, 48, 48, 3) 0
_________________________________________________________________
block1_conv1 (Conv2D) (None, 48, 48, 64) 1792
_________________________________________________________________
block1_conv2 (Conv2D) (None, 48, 48, 64) 36928
_________________________________________________________________
block1_pool (MaxPooling2D) (None, 24, 24, 64) 0
_________________________________________________________________
block2_conv1 (Conv2D) (None, 24, 24, 128) 73856
_________________________________________________________________
block2_conv2 (Conv2D) (None, 24, 24, 128) 147584
_________________________________________________________________
block2_pool (MaxPooling2D) (None, 12, 12, 128) 0
_________________________________________________________________
block3_conv1 (Conv2D) (None, 12, 12, 256) 295168
_________________________________________________________________
block3_conv2 (Conv2D) (None, 12, 12, 256) 590080
_________________________________________________________________
block3_conv3 (Conv2D) (None, 12, 12, 256) 590080
_________________________________________________________________
block3_conv4 (Conv2D) (None, 12, 12, 256) 590080
_________________________________________________________________
block3_pool (MaxPooling2D) (None, 6, 6, 256) 0
_________________________________________________________________
block4_conv1 (Conv2D) (None, 6, 6, 512) 1180160
_________________________________________________________________
block4_conv2 (Conv2D) (None, 6, 6, 512) 2359808
_________________________________________________________________
block4_conv3 (Conv2D) (None, 6, 6, 512) 2359808
_________________________________________________________________
block4_conv4 (Conv2D) (None, 6, 6, 512) 2359808
_________________________________________________________________
block4_pool (MaxPooling2D) (None, 3, 3, 512) 0
_________________________________________________________________
block5_conv1 (Conv2D) (None, 3, 3, 512) 2359808
_________________________________________________________________
block5_conv2 (Conv2D) (None, 3, 3, 512) 2359808
_________________________________________________________________
block5_conv3 (Conv2D) (None, 3, 3, 512) 2359808
_________________________________________________________________
block5_conv4 (Conv2D) (None, 3, 3, 512) 2359808
_________________________________________________________________
block5_pool (MaxPooling2D) (None, 1, 1, 512) 0
_________________________________________________________________
flatten (Flatten) (None, 512) 0
_________________________________________________________________
fc1 (Dense) (None, 4096) 2101248
_________________________________________________________________
fc2 (Dense) (None, 4096) 16781312
_________________________________________________________________
predictions (Dense) (None, 100) 409700
=================================================================
Total params: 39,316,644
Trainable params: 39,316,644
Non-trainable params: 0
De 34 millones a 39. Ahora sólo queda entrenar.
vgg19 = vgg19_model.fit(x=x_train_resized, y=y_train, batch_size=32, epochs=10, verbose=1, validation_data=(x_test_resized, y_test), shuffle=True)
Train on 50000 samples, validate on 10000 samples
Epoch 1/10
50000/50000 [==============================] - 208s 4ms/step - loss: 4.6054 - acc: 0.0085 - mean_squared_error: 0.0099 - val_loss: 4.6051 - val_acc: 0.0113 - val_mean_squared_error: 0.0099
Epoch 2/10
50000/50000 [==============================] - 207s 4ms/step - loss: 4.6053 - acc: 0.0094 - mean_squared_error: 0.0099 - val_loss: 4.6051 - val_acc: 0.0100 - val_mean_squared_error: 0.0099
Epoch 3/10
50000/50000 [==============================] - 207s 4ms/step - loss: 4.6053 - acc: 0.0097 - mean_squared_error: 0.0099 - val_loss: 4.6051 - val_acc: 0.0154 - val_mean_squared_error: 0.0099
Epoch 4/10
50000/50000 [==============================] - 206s 4ms/step - loss: 4.6052 - acc: 0.0106 - mean_squared_error: 0.0099 - val_loss: 4.6050 - val_acc: 0.0100 - val_mean_squared_error: 0.0099
Epoch 5/10
50000/50000 [==============================] - 207s 4ms/step - loss: 4.6051 - acc: 0.0117 - mean_squared_error: 0.0099 - val_loss: 4.6047 - val_acc: 0.0152 - val_mean_squared_error: 0.0099
Epoch 6/10
50000/50000 [==============================] - 207s 4ms/step - loss: 4.6046 - acc: 0.0149 - mean_squared_error: 0.0099 - val_loss: 4.6038 - val_acc: 0.0186 - val_mean_squared_error: 0.0099
Epoch 7/10
50000/50000 [==============================] - 207s 4ms/step - loss: 4.6027 - acc: 0.0173 - mean_squared_error: 0.0099 - val_loss: 4.6003 - val_acc: 0.0169 - val_mean_squared_error: 0.0099
Epoch 8/10
50000/50000 [==============================] - 207s 4ms/step - loss: 4.5682 - acc: 0.0182 - mean_squared_error: 0.0099 - val_loss: 4.4942 - val_acc: 0.0185 - val_mean_squared_error: 0.0099
Epoch 9/10
50000/50000 [==============================] - 207s 4ms/step - loss: 4.4183 - acc: 0.0296 - mean_squared_error: 0.0099 - val_loss: 4.3578 - val_acc: 0.0315 - val_mean_squared_error: 0.0098
Epoch 10/10
50000/50000 [==============================] - 207s 4ms/step - loss: 4.3185 - acc: 0.0362 - mean_squared_error: 0.0098 - val_loss: 4.2512 - val_acc: 0.0513 - val_mean_squared_error: 0.0098
Obviaremos la evaluación.
Veamos las métricas obtenidas para el entrenamiento y validación gráficamente.
plt.figure(0)
plt.plot(vgg19.history['acc'],'r')
plt.plot(vgg19.history['val_acc'],'g')
plt.xticks(np.arange(0, 11, 2.0))
plt.rcParams['figure.figsize'] = (8, 6)
plt.xlabel("Num of Epochs")
plt.ylabel("Accuracy")
plt.title("Training Accuracy vs Validation Accuracy")
plt.legend(['train','validation'])
plt.figure(1)
plt.plot(vgg19.history['loss'],'r')
plt.plot(vgg19.history['val_loss'],'g')
plt.xticks(np.arange(0, 11, 2.0))
plt.rcParams['figure.figsize'] = (8, 6)
plt.xlabel("Num of Epochs")
plt.ylabel("Loss")
plt.title("Training Loss vs Validation Loss")
plt.legend(['train','validation'])
plt.show()
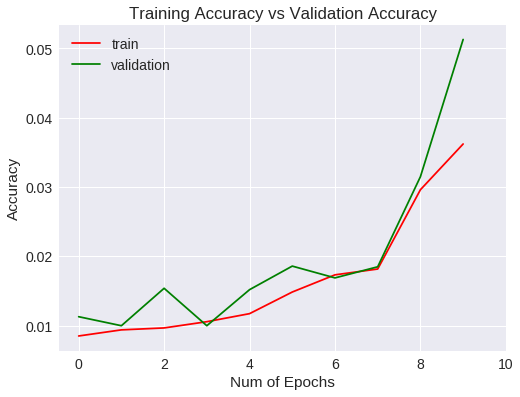
Accuracy
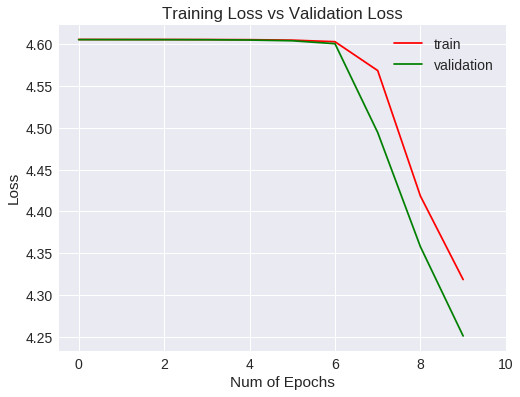
Loss
La generalización mejora a un 1% aproximadamente respecto a la red convolucional del experimento anterior, si bien, después de 10 epochs no ha mejorado las métricas estándar.
Matriz de confusión
Pasemos ahora a ver la matriz de confusión y las métricas de Accuracy, Recall y F1-score.
Vamos a hacer una predicción sobre el dataset de validación y, a partir de ésta, generamos la matriz de confusión y mostramos las métricas mencionadas anteriormente.
vgg19_pred = vgg19_model.predict(x_test_resized, batch_size=32, verbose=1)
vgg19_predicted = np.argmax(vgg19_pred, axis=1)
Como ya hiciéramos en la primera parte, vamos a dar como predecida el mayor valor de la predicción. Lo normal es dar un valor mínimo o bias que defina un resultado como positivo, pero en este caso, lo vamos a hacer simple.
Con la librería Scikit Learn, generamos la matriz de confusión y la dibujamos (aunque el gráfico no es muy bueno debido al numero de etiquetas).
#Creamos la matriz de confusión
vgg19_cm = confusion_matrix(np.argmax(y_test, axis=1), vgg19_predicted)
# Visualiamos la matriz de confusión
vgg19_df_cm = pd.DataFrame(vgg19_cm, range(100), range(100))
plt.figure(figsize = (20,14))
sn.set(font_scale=1.4) #for label size
sn.heatmap(vgg19_df_cm, annot=True, annot_kws={"size": 12}) # font size
plt.show()

Matriz de confusión
Y por último, mostramos las métricas
vgg19_report = classification_report(np.argmax(y_test, axis=1), vgg19_predicted)
print(vgg19_report)
precision recall f1-score support
0 0.00 0.00 0.00 100
1 0.00 0.00 0.00 100
2 0.00 0.00 0.00 100
3 0.00 0.00 0.00 100
4 0.00 0.00 0.00 100
5 0.00 0.00 0.00 100
6 0.00 0.00 0.00 100
7 0.00 0.00 0.00 100
8 0.00 0.00 0.00 100
9 0.00 0.00 0.00 100
10 0.00 0.00 0.00 100
11 0.00 0.00 0.00 100
12 0.00 0.00 0.00 100
13 0.00 0.00 0.00 100
14 0.02 0.01 0.01 100
15 0.00 0.00 0.00 100
16 0.00 0.00 0.00 100
17 0.07 0.08 0.07 100
18 0.00 0.00 0.00 100
19 0.00 0.00 0.00 100
20 0.05 0.55 0.09 100
21 0.11 0.07 0.09 100
22 0.04 0.08 0.06 100
23 0.00 0.00 0.00 100
24 0.08 0.22 0.11 100
25 0.00 0.00 0.00 100
26 0.00 0.00 0.00 100
27 0.00 0.00 0.00 100
28 0.06 0.06 0.06 100
29 0.00 0.00 0.00 100
30 0.07 0.53 0.13 100
31 0.02 0.02 0.02 100
32 0.00 0.00 0.00 100
33 0.00 0.00 0.00 100
34 0.00 0.00 0.00 100
35 0.00 0.00 0.00 100
36 0.01 0.03 0.02 100
37 0.00 0.00 0.00 100
38 0.00 0.00 0.00 100
39 0.07 0.04 0.05 100
40 0.00 0.00 0.00 100
41 0.03 0.04 0.04 100
42 0.00 0.00 0.00 100
43 0.04 0.28 0.08 100
44 0.00 0.00 0.00 100
45 0.00 0.00 0.00 100
46 0.00 0.00 0.00 100
47 0.08 0.25 0.13 100
48 0.00 0.00 0.00 100
49 0.07 0.33 0.12 100
50 0.00 0.00 0.00 100
51 0.00 0.00 0.00 100
52 0.10 0.43 0.16 100
53 0.04 0.79 0.08 100
54 0.00 0.00 0.00 100
55 0.04 0.01 0.02 100
56 0.00 0.00 0.00 100
57 0.00 0.00 0.00 100
58 0.09 0.04 0.06 100
59 0.00 0.00 0.00 100
60 0.23 0.11 0.15 100
61 0.00 0.00 0.00 100
62 0.00 0.00 0.00 100
63 0.00 0.00 0.00 100
64 0.00 0.00 0.00 100
65 0.00 0.00 0.00 100
66 0.00 0.00 0.00 100
67 0.00 0.00 0.00 100
68 0.00 0.00 0.00 100
69 0.00 0.00 0.00 100
70 0.00 0.00 0.00 100
71 0.07 0.10 0.08 100
72 0.00 0.00 0.00 100
73 0.00 0.00 0.00 100
74 0.02 0.01 0.01 100
75 0.05 0.11 0.07 100
76 0.00 0.00 0.00 100
77 0.00 0.00 0.00 100
78 0.00 0.00 0.00 100
79 0.00 0.00 0.00 100
80 0.00 0.00 0.00 100
81 0.04 0.09 0.05 100
82 0.08 0.01 0.02 100
83 0.00 0.00 0.00 100
84 0.00 0.00 0.00 100
85 0.04 0.04 0.04 100
86 0.00 0.00 0.00 100
87 0.00 0.00 0.00 100
88 0.03 0.31 0.05 100
89 0.00 0.00 0.00 100
90 0.00 0.00 0.00 100
91 0.00 0.00 0.00 100
92 0.00 0.00 0.00 100
93 0.00 0.00 0.00 100
94 0.00 0.00 0.00 100
95 0.11 0.25 0.15 100
96 0.00 0.00 0.00 100
97 0.04 0.24 0.07 100
98 0.00 0.00 0.00 100
99 0.00 0.00 0.00 100
avg / total 0.02 0.05 0.02 10000
Curva ROC (tasas de verdaderos positivos y falsos positivos)
Vamos a codificar la curva ROC.
from sklearn.datasets import make_classification
from sklearn.preprocessing import label_binarize
from scipy import interp
from itertools import cycle
n_classes = 100
from sklearn.metrics import roc_curve, auc
# Plot linewidth.
lw = 2
# Compute ROC curve and ROC area for each class
fpr = dict()
tpr = dict()
roc_auc = dict()
for i in range(n_classes):
fpr[i], tpr[i], _ = roc_curve(y_test[:, i], vgg19_pred[:, i])
roc_auc[i] = auc(fpr[i], tpr[i])
# Compute micro-average ROC curve and ROC area
fpr["micro"], tpr["micro"], _ = roc_curve(y_test.ravel(), vgg19_pred.ravel())
roc_auc["micro"] = auc(fpr["micro"], tpr["micro"])
# Compute macro-average ROC curve and ROC area
# First aggregate all false positive rates
all_fpr = np.unique(np.concatenate([fpr[i] for i in range(n_classes)]))
# Then interpolate all ROC curves at this points
mean_tpr = np.zeros_like(all_fpr)
for i in range(n_classes):
mean_tpr += interp(all_fpr, fpr[i], tpr[i])
# Finally average it and compute AUC
mean_tpr /= n_classes
fpr["macro"] = all_fpr
tpr["macro"] = mean_tpr
roc_auc["macro"] = auc(fpr["macro"], tpr["macro"])
# Plot all ROC curves
plt.figure(1)
plt.plot(fpr["micro"], tpr["micro"],
label='micro-average ROC curve (area = {0:0.2f})'
''.format(roc_auc["micro"]),
color='deeppink', linestyle=':', linewidth=4)
plt.plot(fpr["macro"], tpr["macro"],
label='macro-average ROC curve (area = {0:0.2f})'
''.format(roc_auc["macro"]),
color='navy', linestyle=':', linewidth=4)
colors = cycle(['aqua', 'darkorange', 'cornflowerblue'])
for i, color in zip(range(n_classes-97), colors):
plt.plot(fpr[i], tpr[i], color=color, lw=lw,
label='ROC curve of class {0} (area = {1:0.2f})'
''.format(i, roc_auc[i]))
plt.plot([0, 1], [0, 1], 'k--', lw=lw)
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Some extension of Receiver operating characteristic to multi-class')
plt.legend(loc="lower right")
plt.show()
# Zoom in view of the upper left corner.
plt.figure(2)
plt.xlim(0, 0.2)
plt.ylim(0.8, 1)
plt.plot(fpr["micro"], tpr["micro"],
label='micro-average ROC curve (area = {0:0.2f})'
''.format(roc_auc["micro"]),
color='deeppink', linestyle=':', linewidth=4)
plt.plot(fpr["macro"], tpr["macro"],
label='macro-average ROC curve (area = {0:0.2f})'
''.format(roc_auc["macro"]),
color='navy', linestyle=':', linewidth=4)
colors = cycle(['aqua', 'darkorange', 'cornflowerblue'])
for i, color in zip(range(3), colors):
plt.plot(fpr[i], tpr[i], color=color, lw=lw,
label='ROC curve of class {0} (area = {1:0.2f})'
''.format(i, roc_auc[i]))
plt.plot([0, 1], [0, 1], 'k--', lw=lw)
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Some extension of Receiver operating characteristic to multi-class')
plt.legend(loc="lower right")
plt.show()
El resultado para tres clases se muestra en los siguientes gráficos.
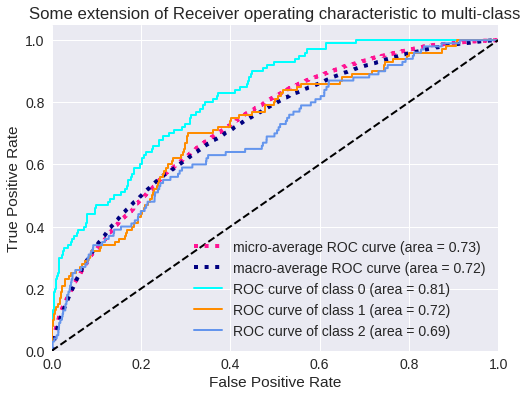
Curva ROC para 3 clases
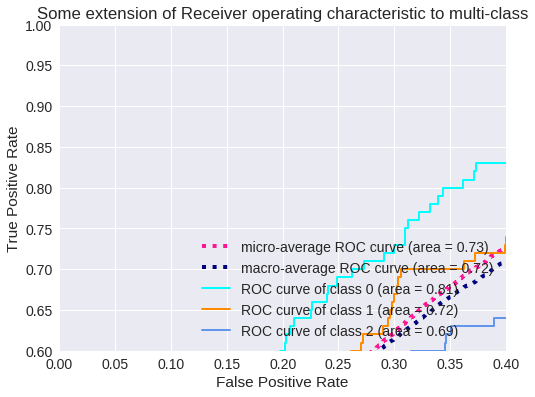
Zoom de la Curva ROC para 3 clases
Vamos a ver algunos resultados
imgplot = plt.imshow(x_train_original[0])
plt.show()
print('class for image 1: ' + str(np.argmax(y_test[0])))
print('predicted: ' + str(vgg19_predicted[0]))

Una vaca?
class for image 1: 49
predicted: 30
imgplot = plt.imshow(x_train_original[3])
plt.show()
print('class for image 3: ' + str(np.argmax(y_test[3])))
print('predicted: ' + str(vgg19_predicted[3]))

Un hombre?
class for image 3: 51
predicted: 75
Salvaremos los datos del histórico de entrenamiento para compararlos con otros modelos.
#Histórico
with open(path_base + '/vgg19_history.txt', 'wb') as file_pi:
pickle.dump(scnn.history, file_pi)
A continuación, vamos a comparar las métricas con los modelos anteriores.
with open(path_base + '/vgg16_history.txt', 'rb') as f:
vgg16_history = pickle.load(f)
Ya lo tenemos en las variables correspondientes. Ahora, comparemos las gráficas.
plt.figure(0)
plt.plot(snn_history['val_acc'],'r')
plt.plot(scnn_history['val_acc'],'g')
plt.plot(vgg16_history['val_acc'],'b')
plt.plot(vgg19.history['val_acc'],'y')
plt.xticks(np.arange(0, 11, 2.0))
plt.rcParams['figure.figsize'] = (8, 6)
plt.xlabel("Num of Epochs")
plt.ylabel("Accuracy")
plt.title("Simple NN Accuracy vs simple CNN Accuracy")
plt.legend(['simple NN','CNN','VGG 16','VGG 19'])
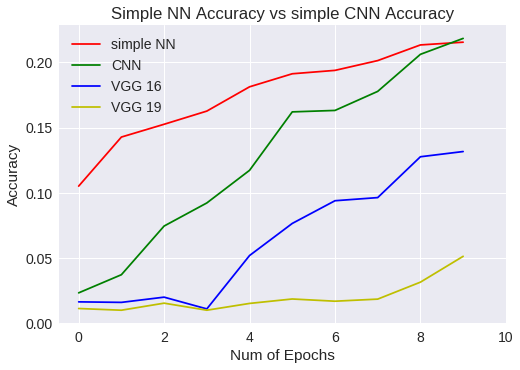
Simple NN Vs CNN accuracy
plt.figure(0)
plt.plot(snn_history['val_loss'],'r')
plt.plot(scnn_history['val_loss'],'g')
plt.plot(vgg16.history['val_loss'],'b')
plt.plot(vgg19.history['val_loss'],'y')
plt.xticks(np.arange(0, 11, 2.0))
plt.rcParams['figure.figsize'] = (8, 6)
plt.xlabel("Num of Epochs")
plt.ylabel("Loss")
plt.title("Simple NN Loss vs simple CNN Loss")
plt.legend(['simple NN','CNN','VGG 16','VGG 19'])
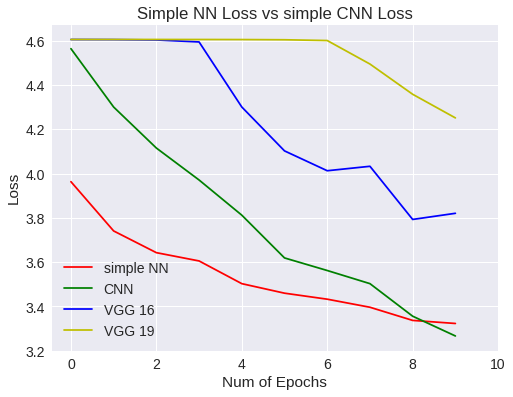
Simple NN Vs CNN loss
plt.figure(0)
plt.plot(snn_history['val_mean_squared_error'],'r')
plt.plot(scnn_history['val_mean_squared_error'],'g')
plt.plot(vgg16.history['val_mean_squared_error'],'b')
plt.plot(vgg19.history['val_mean_squared_error'],'y')
plt.xticks(np.arange(0, 11, 2.0))
plt.rcParams['figure.figsize'] = (8, 6)
plt.xlabel("Num of Epochs")
plt.ylabel("Mean Squared Error")
plt.title("Simple NN MSE vs simple CNN MSE")
plt.legend(['simple NN','CNN','VGG 16','VGG 19'])
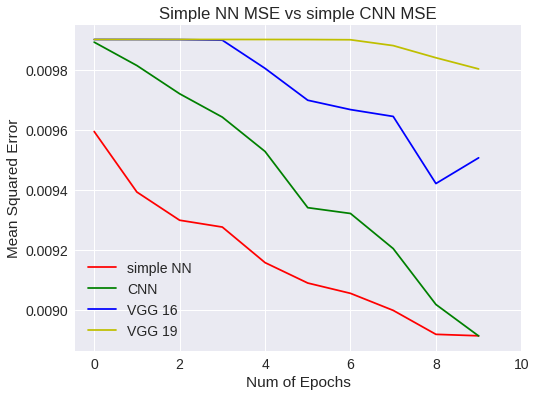
Simple NN Vs CNN MSE
¿Que ha pasado?
Bien, la respuesta es sencilla. Hemos querido usar un modelo predeterminado, obligándonos a modificar la imagen original haciéndola más grande. Como la imagen es de 32x32 píxeles, pues la imagen resultante queda peor y aprende peor. Esto es, hemos cambiado el ámbito de los datos con respecto a los experimentos anteriores.
Así que lo que tenemos que hacer, es una red VGG que no necesite modificar la imagen original.
Custom VGG
Definamos el modelo más específicamente.
def VGG16_Without_lastPool(include_top=True, input_tensor=None, input_shape=(32,32,3), pooling=None, classes=100):
if input_tensor is None:
img_input = Input(shape=input_shape)
else:
if not K.is_keras_tensor(input_tensor):
img_input = Input(tensor=input_tensor, shape=input_shape)
else:
img_input = input_tensor
# Block 1
x = Conv2D(64, (3, 3), activation='relu', padding='same', name='block1_conv1')(img_input)
x = Conv2D(64, (3, 3), activation='relu', padding='same', name='block1_conv2')(x)
x = MaxPooling2D((2, 2), strides=(2, 2), name='block1_pool')(x) #to 16x16
# Block 2
x = Conv2D(128, (3, 3), activation='relu', padding='same', name='block2_conv1')(x)
x = Conv2D(128, (3, 3), activation='relu', padding='same', name='block2_conv2')(x)
x = MaxPooling2D((2, 2), strides=(2, 2), name='block2_pool')(x) #to 8x8
# Block 3
x = Conv2D(256, (3, 3), activation='relu', padding='same', name='block3_conv1')(x)
x = Conv2D(256, (3, 3), activation='relu', padding='same', name='block3_conv2')(x)
x = Conv2D(256, (3, 3), activation='relu', padding='same', name='block3_conv3')(x)
x = MaxPooling2D((2, 2), strides=(2, 2), name='block3_pool')(x) #to 4x4
# Block 4
x = Conv2D(512, (3, 3), activation='relu', padding='same', name='block4_conv1')(x)
x = Conv2D(512, (3, 3), activation='relu', padding='same', name='block4_conv2')(x)
x = Conv2D(512, (3, 3), activation='relu', padding='same', name='block4_conv3')(x)
x = MaxPooling2D((2, 2), strides=(2, 2), name='block4_pool')(x) #to 2x2
# Block 5
x = Conv2D(512, (3, 3), activation='relu', padding='same', name='block5_conv1')(x)
x = Conv2D(512, (3, 3), activation='relu', padding='same', name='block5_conv2')(x)
x = Conv2D(512, (3, 3), activation='relu', padding='same', name='block5_conv3')(x)
#x = MaxPooling2D((2, 2), strides=(2, 2), name='block5_pool')(x)
if include_top:
# Classification block
x = Flatten(name='flatten')(x)
x = Dense(4096, activation='relu', name='fc1')(x)
x = Dense(4096, activation='relu', name='fc2')(x)
x = Dense(classes, activation='softmax', name='predictions')(x)
else:
if pooling == 'avg':
x = GlobalAveragePooling2D()(x)
elif pooling == 'max':
x = GlobalMaxPooling2D()(x)
# Create model.
model = Model(img_input, x, name='vgg16Bis')
return model
Compilamos como hasta ahora...
def create_vgg16WithoutPool():
model = VGG16_Without_lastPool(include_top=True, input_tensor=None, input_shape=(32,32,3), pooling=None, classes=100)
return model
vgg16Bis_model = create_vgg16WithoutPool()
vgg16Bis_model.compile(loss='categorical_crossentropy', optimizer='sgd', metrics=['acc', 'mse'])
Una vez hecho esto, vamos a ver un resumen del modelo creado.
vgg16Bis_model.summary()
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input_3 (InputLayer) (None, 32, 32, 3) 0
_________________________________________________________________
block1_conv1 (Conv2D) (None, 32, 32, 64) 1792
_________________________________________________________________
block1_conv2 (Conv2D) (None, 32, 32, 64) 36928
_________________________________________________________________
block1_pool (MaxPooling2D) (None, 16, 16, 64) 0
_________________________________________________________________
block2_conv1 (Conv2D) (None, 16, 16, 128) 73856
_________________________________________________________________
block2_conv2 (Conv2D) (None, 16, 16, 128) 147584
_________________________________________________________________
block2_pool (MaxPooling2D) (None, 8, 8, 128) 0
_________________________________________________________________
block3_conv1 (Conv2D) (None, 8, 8, 256) 295168
_________________________________________________________________
block3_conv2 (Conv2D) (None, 8, 8, 256) 590080
_________________________________________________________________
block3_conv3 (Conv2D) (None, 8, 8, 256) 590080
_________________________________________________________________
block3_pool (MaxPooling2D) (None, 4, 4, 256) 0
_________________________________________________________________
block4_conv1 (Conv2D) (None, 4, 4, 512) 1180160
_________________________________________________________________
block4_conv2 (Conv2D) (None, 4, 4, 512) 2359808
_________________________________________________________________
block4_conv3 (Conv2D) (None, 4, 4, 512) 2359808
_________________________________________________________________
block4_pool (MaxPooling2D) (None, 2, 2, 512) 0
_________________________________________________________________
block5_conv1 (Conv2D) (None, 2, 2, 512) 2359808
_________________________________________________________________
block5_conv2 (Conv2D) (None, 2, 2, 512) 2359808
_________________________________________________________________
block5_conv3 (Conv2D) (None, 2, 2, 512) 2359808
_________________________________________________________________
flatten (Flatten) (None, 2048) 0
_________________________________________________________________
fc1 (Dense) (None, 4096) 8392704
_________________________________________________________________
fc2 (Dense) (None, 4096) 16781312
_________________________________________________________________
predictions (Dense) (None, 100) 409700
=================================================================
Total params: 40,298,404
Trainable params: 40,298,404
Non-trainable params: 0
Tenemos uno más, 40 millones de parámetros, porque en realizad, sólo hemos quitado el Pool del bloque 5. Ahora sólo queda entrenar, pero esta vez con el dataset original normalizado.
vgg16Bis = vgg16Bis_model.fit(x=x_train, y=y_train, batch_size=32, epochs=10, verbose=1, validation_data=(x_test, y_test), shuffle=True)
Train on 50000 samples, validate on 10000 samples
Epoch 1/10
50000/50000 [==============================] - 139s 3ms/step - loss: 4.6053 - acc: 0.0093 - mean_squared_error: 0.0099 - val_loss: 4.6051 - val_acc: 0.0100 - val_mean_squared_error: 0.0099
Epoch 2/10
50000/50000 [==============================] - 137s 3ms/step - loss: 4.6053 - acc: 0.0100 - mean_squared_error: 0.0099 - val_loss: 4.6050 - val_acc: 0.0099 - val_mean_squared_error: 0.0099
Epoch 3/10
50000/50000 [==============================] - 137s 3ms/step - loss: 4.6051 - acc: 0.0105 - mean_squared_error: 0.0099 - val_loss: 4.6048 - val_acc: 0.0174 - val_mean_squared_error: 0.0099
Epoch 4/10
50000/50000 [==============================] - 138s 3ms/step - loss: 4.6048 - acc: 0.0119 - mean_squared_error: 0.0099 - val_loss: 4.6042 - val_acc: 0.0198 - val_mean_squared_error: 0.0099
Epoch 5/10
50000/50000 [==============================] - 137s 3ms/step - loss: 4.6036 - acc: 0.0210 - mean_squared_error: 0.0099 - val_loss: 4.6020 - val_acc: 0.0228 - val_mean_squared_error: 0.0099
Epoch 6/10
50000/50000 [==============================] - 137s 3ms/step - loss: 4.5910 - acc: 0.0231 - mean_squared_error: 0.0099 - val_loss: 4.4990 - val_acc: 0.0203 - val_mean_squared_error: 0.0099
Epoch 7/10
50000/50000 [==============================] - 138s 3ms/step - loss: 4.4632 - acc: 0.0217 - mean_squared_error: 0.0099 - val_loss: 4.4322 - val_acc: 0.0227 - val_mean_squared_error: 0.0099
Epoch 8/10
50000/50000 [==============================] - 138s 3ms/step - loss: 4.3865 - acc: 0.0261 - mean_squared_error: 0.0098 - val_loss: 4.3199 - val_acc: 0.0315 - val_mean_squared_error: 0.0098
Epoch 9/10
50000/50000 [==============================] - 137s 3ms/step - loss: 4.2913 - acc: 0.0300 - mean_squared_error: 0.0098 - val_loss: 4.2396 - val_acc: 0.0348 - val_mean_squared_error: 0.0098
Epoch 10/10
50000/50000 [==============================] - 138s 3ms/step - loss: 4.2151 - acc: 0.0363 - mean_squared_error: 0.0098 - val_loss: 4.3424 - val_acc: 0.0256 - val_mean_squared_error: 0.0098
Veamos las métricas obtenidas para el entrenamiento y validación gráficamente.
plt.figure(0)
plt.plot(vgg16Bis.history['acc'],'r')
plt.plot(vgg16Bis.history['val_acc'],'g')
plt.xticks(np.arange(0, 11, 2.0))
plt.rcParams['figure.figsize'] = (8, 6)
plt.xlabel("Num of Epochs")
plt.ylabel("Accuracy")
plt.title("Training Accuracy vs Validation Accuracy")
plt.legend(['train','validation'])
plt.figure(1)
plt.plot(vgg16Bis.history['loss'],'r')
plt.plot(vgg16Bis.history['val_loss'],'g')
plt.xticks(np.arange(0, 11, 2.0))
plt.rcParams['figure.figsize'] = (8, 6)
plt.xlabel("Num of Epochs")
plt.ylabel("Loss")
plt.title("Training Loss vs Validation Loss")
plt.legend(['train','validation'])
plt.show()
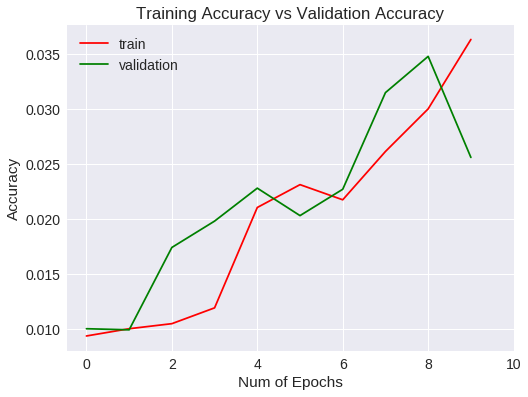
Accuracy
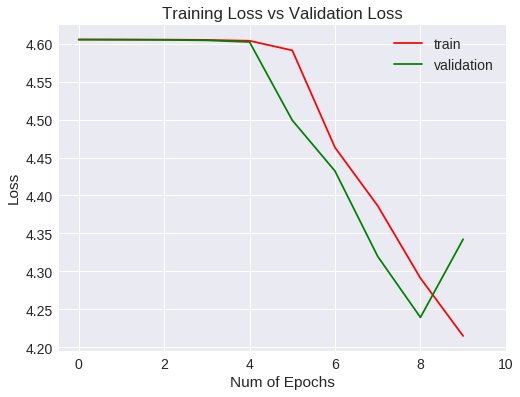
Loss
Después de 10 epochs no ha mejorado las métricas estándar.
Matriz de confusión
Pasemos ahora a ver la matriz de confusión y las métricas de Accuracy, Recall y F1-score.
Vamos a hacer una predicción sobre el dataset de validación y, a partir de ésta, generamos la matriz de confusión y mostramos las métricas mencionadas anteriormente.
vgg16Bis_pred = vgg16Bis_model.predict(x_test, batch_size=32, verbose=1)
vgg16Bis_predicted = np.argmax(vgg16Bis_pred, axis=1)
vgg_cm = confusion_matrix(np.argmax(y_test, axis=1), vgg16Bis_predicted)
# Visualizing of confusion matrix
vgg_df_cm = pd.DataFrame(vgg_cm, range(100), range(100))
plt.figure(figsize = (20,14))
sn.set(font_scale=1.4) #for label size
sn.heatmap(vgg_df_cm, annot=True, annot_kws={"size": 12}) # font size
plt.show()
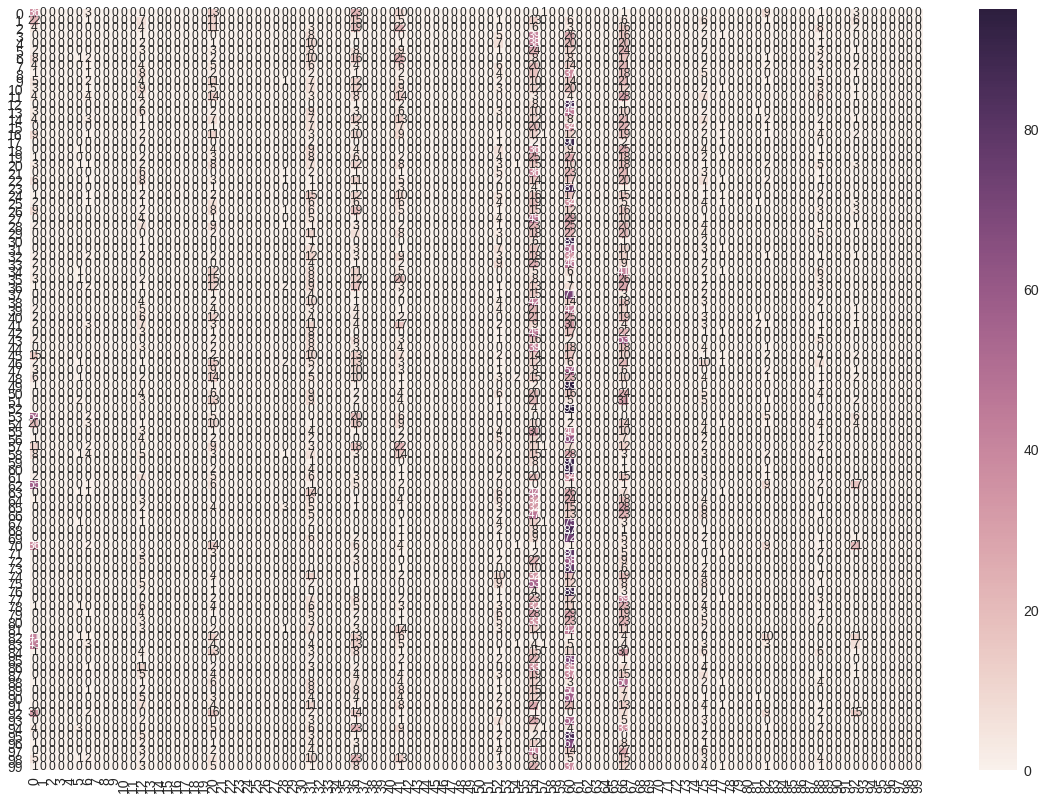
Matriz de confusión
Y por último, mostramos las métricas
vgg_report = classification_report(np.argmax(y_test, axis=1), vgg16Bis_predicted)
print(vgg_report)
precision recall f1-score support
0 0.32 0.34 0.33 100
1 0.19 0.22 0.20 100
2 0.23 0.14 0.17 100
3 0.11 0.02 0.03 100
4 0.03 0.02 0.02 100
5 0.13 0.08 0.10 100
6 0.21 0.19 0.20 100
7 0.13 0.17 0.15 100
8 0.11 0.10 0.10 100
9 0.14 0.24 0.18 100
10 0.00 0.00 0.00 100
11 0.09 0.14 0.11 100
12 0.18 0.02 0.04 100
13 0.14 0.02 0.04 100
14 0.25 0.08 0.12 100
15 0.10 0.05 0.07 100
16 0.31 0.11 0.16 100
17 0.15 0.42 0.22 100
18 0.16 0.32 0.21 100
19 0.10 0.05 0.07 100
20 0.28 0.30 0.29 100
21 0.08 0.29 0.13 100
22 0.00 0.00 0.00 100
23 0.18 0.46 0.26 100
24 0.25 0.30 0.27 100
25 0.06 0.01 0.02 100
26 0.04 0.01 0.02 100
27 0.07 0.08 0.07 100
28 0.28 0.24 0.26 100
29 0.23 0.08 0.12 100
30 0.17 0.12 0.14 100
31 0.07 0.11 0.09 100
32 0.19 0.03 0.05 100
33 0.20 0.12 0.15 100
34 0.12 0.19 0.15 100
35 0.15 0.07 0.10 100
36 0.12 0.04 0.06 100
37 0.10 0.10 0.10 100
38 0.09 0.08 0.08 100
39 0.00 0.00 0.00 100
40 0.17 0.03 0.05 100
41 0.28 0.51 0.36 100
42 0.12 0.13 0.12 100
43 0.18 0.30 0.23 100
44 0.17 0.02 0.04 100
45 0.00 0.00 0.00 100
46 0.18 0.07 0.10 100
47 0.20 0.50 0.28 100
48 0.30 0.11 0.16 100
49 0.20 0.13 0.16 100
50 0.00 0.00 0.00 100
51 0.10 0.09 0.10 100
52 0.32 0.26 0.29 100
53 0.59 0.13 0.21 100
54 0.14 0.53 0.23 100
55 0.00 0.00 0.00 100
56 0.14 0.32 0.19 100
57 0.37 0.10 0.16 100
58 0.13 0.14 0.14 100
59 0.20 0.03 0.05 100
60 0.37 0.71 0.49 100
61 0.13 0.20 0.16 100
62 0.20 0.57 0.30 100
63 0.10 0.28 0.15 100
64 0.00 0.00 0.00 100
65 0.16 0.06 0.09 100
66 0.10 0.01 0.02 100
67 0.16 0.05 0.08 100
68 0.36 0.47 0.41 100
69 0.16 0.29 0.21 100
70 0.32 0.08 0.13 100
71 0.25 0.35 0.29 100
72 0.00 0.00 0.00 100
73 0.20 0.50 0.28 100
74 0.05 0.12 0.07 100
75 0.08 0.30 0.12 100
76 0.17 0.55 0.26 100
77 0.08 0.01 0.02 100
78 0.22 0.02 0.04 100
79 0.15 0.02 0.04 100
80 0.08 0.05 0.06 100
81 0.11 0.10 0.11 100
82 0.59 0.43 0.50 100
83 0.06 0.01 0.02 100
84 0.09 0.06 0.07 100
85 0.19 0.12 0.15 100
86 0.13 0.37 0.19 100
87 0.09 0.08 0.08 100
88 0.01 0.01 0.01 100
89 0.12 0.08 0.09 100
90 0.15 0.12 0.13 100
91 0.25 0.37 0.30 100
92 0.18 0.10 0.13 100
93 0.13 0.11 0.12 100
94 0.18 0.26 0.21 100
95 0.23 0.16 0.19 100
96 0.12 0.06 0.08 100
97 0.08 0.07 0.08 100
98 0.05 0.04 0.04 100
99 0.18 0.19 0.19 100
avg / total 0.16 0.16 0.14 10000
Curva ROC (tasas de verdaderos positivos y falsos positivos)
Vamos a codificar la curva ROC.
from sklearn.datasets import make_classification
from sklearn.preprocessing import label_binarize
from scipy import interp
from itertools import cycle
n_classes = 100
from sklearn.metrics import roc_curve, auc
# Plot linewidth.
lw = 2
# Compute ROC curve and ROC area for each class
fpr = dict()
tpr = dict()
roc_auc = dict()
for i in range(n_classes):
fpr[i], tpr[i], _ = roc_curve(y_test[:, i], vgg16Bis_pred[:, i])
roc_auc[i] = auc(fpr[i], tpr[i])
# Compute micro-average ROC curve and ROC area
fpr["micro"], tpr["micro"], _ = roc_curve(y_test.ravel(), vgg16Bis_pred.ravel())
roc_auc["micro"] = auc(fpr["micro"], tpr["micro"])
# Compute macro-average ROC curve and ROC area
# First aggregate all false positive rates
all_fpr = np.unique(np.concatenate([fpr[i] for i in range(n_classes)]))
# Then interpolate all ROC curves at this points
mean_tpr = np.zeros_like(all_fpr)
for i in range(n_classes):
mean_tpr += interp(all_fpr, fpr[i], tpr[i])
# Finally average it and compute AUC
mean_tpr /= n_classes
fpr["macro"] = all_fpr
tpr["macro"] = mean_tpr
roc_auc["macro"] = auc(fpr["macro"], tpr["macro"])
# Plot all ROC curves
plt.figure(1)
plt.plot(fpr["micro"], tpr["micro"],
label='micro-average ROC curve (area = {0:0.2f})'
''.format(roc_auc["micro"]),
color='deeppink', linestyle=':', linewidth=4)
plt.plot(fpr["macro"], tpr["macro"],
label='macro-average ROC curve (area = {0:0.2f})'
''.format(roc_auc["macro"]),
color='navy', linestyle=':', linewidth=4)
colors = cycle(['aqua', 'darkorange', 'cornflowerblue'])
for i, color in zip(range(n_classes-97), colors):
plt.plot(fpr[i], tpr[i], color=color, lw=lw,
label='ROC curve of class {0} (area = {1:0.2f})'
''.format(i, roc_auc[i]))
plt.plot([0, 1], [0, 1], 'k--', lw=lw)
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Some extension of Receiver operating characteristic to multi-class')
plt.legend(loc="lower right")
plt.show()
# Zoom in view of the upper left corner.
plt.figure(2)
plt.xlim(0, 0.2)
plt.ylim(0.8, 1)
plt.plot(fpr["micro"], tpr["micro"],
label='micro-average ROC curve (area = {0:0.2f})'
''.format(roc_auc["micro"]),
color='deeppink', linestyle=':', linewidth=4)
plt.plot(fpr["macro"], tpr["macro"],
label='macro-average ROC curve (area = {0:0.2f})'
''.format(roc_auc["macro"]),
color='navy', linestyle=':', linewidth=4)
colors = cycle(['aqua', 'darkorange', 'cornflowerblue'])
for i, color in zip(range(3), colors):
plt.plot(fpr[i], tpr[i], color=color, lw=lw,
label='ROC curve of class {0} (area = {1:0.2f})'
''.format(i, roc_auc[i]))
plt.plot([0, 1], [0, 1], 'k--', lw=lw)
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Some extension of Receiver operating characteristic to multi-class')
plt.legend(loc="lower right")
plt.show()
El resultado para tres clases se muestra en los siguientes gráficos.
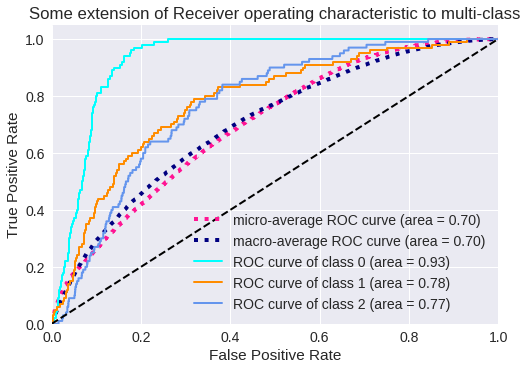
Curva ROC para 3 clases
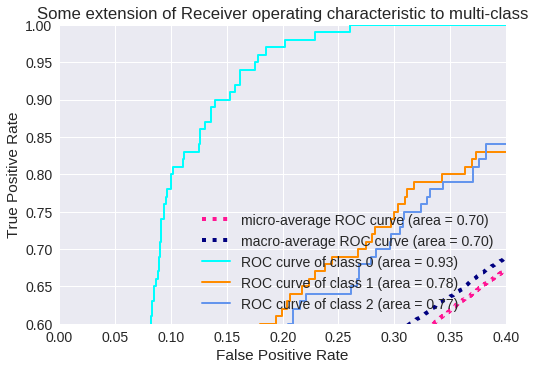
Zoom de la Curva ROC para 3 clases
Salvaremos los datos del histórico de entrenamiento para compararlos con otros modelos.
#Histórico
with open(path_base + '/cvgg16_history.txt', 'wb') as file_pi:
pickle.dump(scnn.history, file_pi)
A continuación, vamos a comparar las métricas con los modelos anteriores.
plt.figure(0)
plt.plot(snn_history['val_acc'],'r')
plt.plot(scnn_history['val_acc'],'g')
plt.plot(vgg16.history['val_acc'],'b')
plt.plot(vgg19.history['val_acc'],'y')
plt.plot(vgg16Bis.history['val_acc'],'m')
plt.xticks(np.arange(0, 11, 2.0))
plt.rcParams['figure.figsize'] = (8, 6)
plt.xlabel("Num of Epochs")
plt.ylabel("Accuracy")
plt.title("Simple NN Accuracy vs simple CNN Accuracy")
plt.legend(['simple NN','CNN','VGG 16','VGG 19','Custom VGG'])
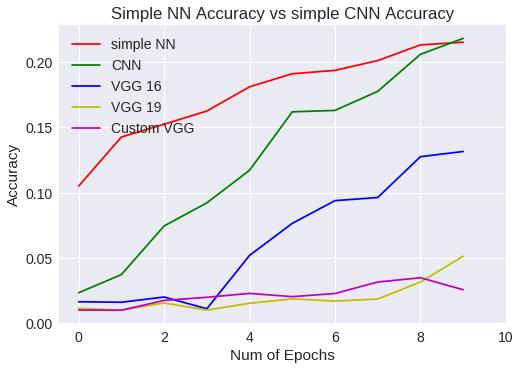
Simple NN Vs CNN accuracy
plt.figure(0)
plt.plot(snn_history['val_loss'],'r')
plt.plot(scnn_history['val_loss'],'g')
plt.plot(vgg16.history['val_loss'],'b')
plt.plot(vgg19.history['val_loss'],'y')
plt.plot(vgg16Bis.history['val_loss'],'m')
plt.xticks(np.arange(0, 11, 2.0))
plt.rcParams['figure.figsize'] = (8, 6)
plt.xlabel("Num of Epochs")
plt.ylabel("Loss")
plt.title("Simple NN Loss vs simple CNN Loss")
plt.legend(['simple NN','CNN','VGG 16','VGG 19','Custom VGG'])
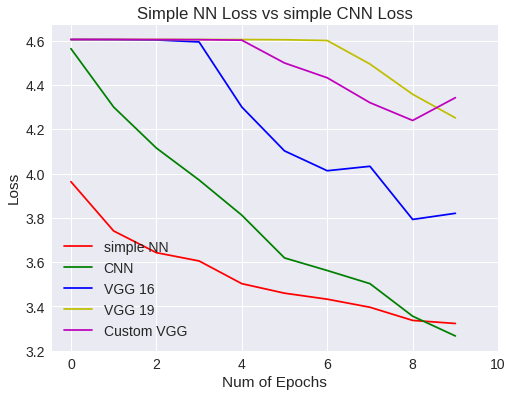
Simple NN Vs CNN loss
plt.figure(0)
plt.plot(snn_history['val_mean_squared_error'],'r')
plt.plot(scnn_history['val_mean_squared_error'],'g')
plt.plot(vgg16.history['val_mean_squared_error'],'b')
plt.plot(vgg19.history['val_mean_squared_error'],'y')
plt.plot(vgg16Bis.history['val_mean_squared_error'],'m')
plt.xticks(np.arange(0, 11, 2.0))
plt.rcParams['figure.figsize'] = (8, 6)
plt.xlabel("Num of Epochs")
plt.ylabel("Mean Squared Error")
plt.title("Simple NN MSE vs simple CNN MSE")
plt.legend(['simple NN','CNN','VGG 16','VGG 19','Custom VGG'])
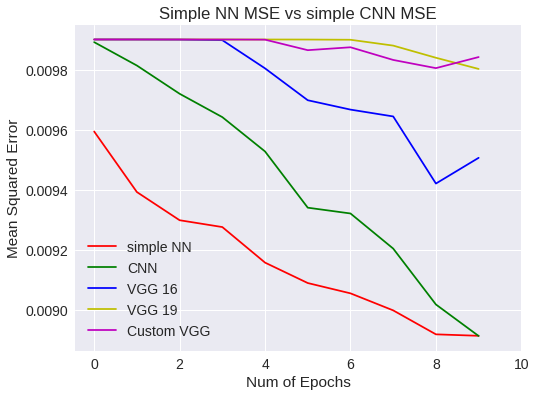
Simple NN Vs CNN MSE
Conclusión sobre el segundo experimento
Hemos visto unos modelos más profundos, en los que el número de parámetros ha aumentado en exceso incluso para una red convolutiva. Además, el aprendizaje es mucho más lento en este caso específico. Quizás, a la vista de los resultados, sería planteable aumentar el número de epochs a realizar, pero en cambio, sabemos que actualmente existen modelos mejores que vamos a ver.
En el siguiente artículo, presentaremos la arquitectura: ResNet.
 Jesús Utrera Burgal
Jesús Utrera Burgal